ASAP is supporting iNDI-PD (an expansion to the original iPSC Neurodegenerative Disease Initiative (iNDI) and anchored within the NIH Center for Alzheimer’s and Related Dementias (CARD)) to make iPSC resources accessible to all researchers interested in understanding Parkinson’s disease (PD) to discover novel insights into the molecular biology of disease and to ensure that additional mutations relevant to PD research are also included in their efforts.
iNDI-PD has added new cell lines associated with PD variants to the Human iPSC catalog through The Jackson Laboratory (JAX). These cell line trios are publicly available for the research community to attribute observed changes to specific mutations more quickly – cutting through the noise of genetic variation – and more confidently.
Induced pluripotent stem cells (iPSCs) are derived from adult cells such as skin or blood and reprogrammed back into pluripotent stem cells. These appealing research tools create the ability to study a disorder in disease-relevant cell types from real patients. However, donors who provide these cells carry innumerable genetic differences, making it hard to pinpoint whether an observation is due to a disease-causing mutation or something else in the patient’s genetic makeup. By incorporating PD data in the neurodegenerative resource bank, we are also enabling researchers to draw connections across diagnostic lines.
iNDI introduces disease-causing mutations to a set of cell lines that all have the same genetic background (called isogenic lines). iNDI is led by Mark Cookson, Ph.D., of the National Institute on Aging, and Michael E. Ward, M.D., Ph.D., of the National Institute of Neurological Disorders and Stroke, both parts of the National Institutes of Health. The initiative was initially funded to model more than 100 mutations associated with Alzheimer’s disease and related dementias in isogenic iPSC lines.
“iNDI is the largest iPSC genome engineering initiative in research to date. The expansion of iNDI to include additional PD lines will allow us not only to discover novel insights into the molecular biology of disease but also to draw connections across diagnostic lines.”
–Mark Cookson, PhD, iNDI Principal Investigator
Overview of iNDI
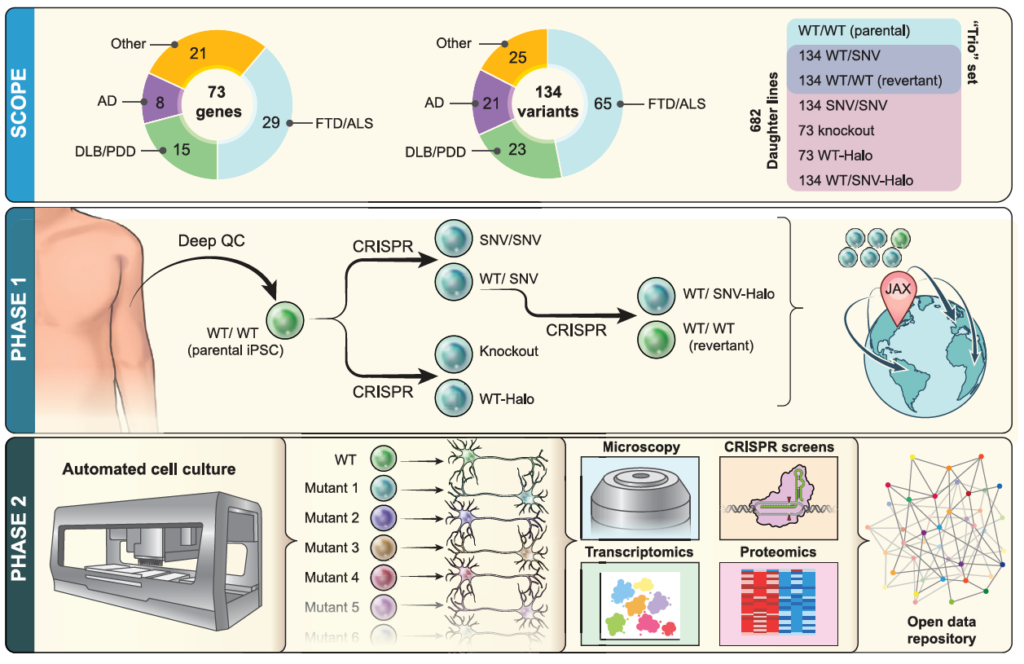
iNDI is a large iPSC genome-engineering initiative, with more than 600 engineered daughter lines to be generated from each parental line. Companion lines will include revertants, Halo-tagged knockins, and knockouts.
[Courtesy of Ramos et al., Neuron, 2021.]
Phase 1:
iNDI will genetically engineer isogenic iPSC lines harboring many ADRD variants, knockouts, and endogenous tags across several deeply characterized parental lines from unaffected individuals. Following stringent quality control assays, each line will be available for distribution through The Jackson Laboratory (JAX).
Phase 2:
Using high-throughput robotic platforms, iNDI will differentiate mutation harboring iPSC lines into CNS-relevant cell types, such as neurons and microglia, followed by a series of phenotypic assays. These include transcriptomics, proteomics, imaging, and genetic interaction screens, yielding a rich multi-dimensional picture of how mutations disrupt cellular function. Open access datasets will enable the community to analyze these data in new ways.
Parkinson's Genes
iNDI already has some PD gene mutations of interest that are associated with dementia within its existing workflow.
Using CRISPR/Cas-mediated gene editing, iNDI-PD will:
- Include a series of human isogenic iPSC lines expressing PD-associated mutations of interest that do not have dementia as a clinical phenotype. (See table for the list of potential mutations that the team is considering)
- Create isogenic controls for genetic PD patient-derived iPSC lines (LRRK2p.G2019S, LRRK2p.R144G, GBA p.N370S, SNCA p.A53T) from the MJFF PPMI cohort
Genes
Mutations of Interest
SNCA
A30P
A53T
E46K
LRRK2
I2020T
N2081D
R1441G
R1441C
R1441H
Y1699C
PINK1
A217D
P399L
PRKN
P437L
R275W
R42P
T240M

